Diffusion mechanisms of DNA in agarose gels: NMR Studies and Monte Carlo Simulations
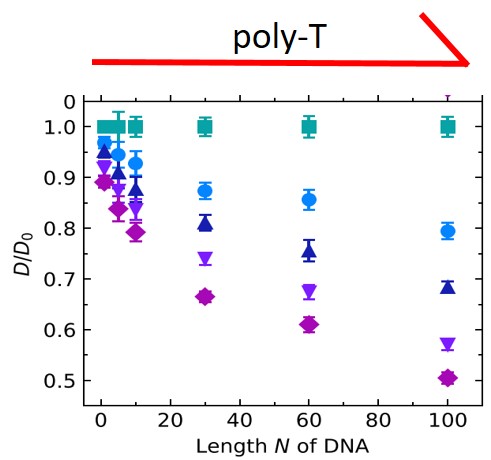
We report on the diffusion mechanism of short, single-stranded DNA molecules with up to 100 nucleobases in agarose gels with concentrations of up to 2.0% with the aim to characterize the DNA–agarose interaction. The diffusion coefficients were measured directly, i.e., without any model assumptions, by pulsed field gradient nuclear magnetic resonance (PFG-NMR). We find that the diffusion coefficient decreases, as expected, with an increase in both DNA strand length and gel concentration. In addition, we performed Monte Carlo simulations of particle diffusion in a model network of polymer chains, considering our experimental conditions. Together, the Monte Carlo simulations and the PFG-NMR results show that the decrease in diffusion coefficients in the presence of the agarose gel is due to a temporary adhesion of the DNA molecules to the surface of gel fibers. The average adhesion time to a given gel fiber increases with the length of the DNA strands but is independent of the number of gel fibers. The corresponding magnitude of the binding enthalpies of DNA strands to gel fibers indicates that a mixture of van der Waals interactions and hydrogen bonding contributes to the decreased diffusion of DNA in agarose gels.
| Author(s): | Bochert, Ida and Günther, Jan-Philipp and Fischer, Peer and Majer, Günter |
| Journal: | The Journal of Chemical Physics |
| Volume: | 156 |
| Number (issue): | 24 |
| Pages: | 245103 |
| Year: | 2022 |
| Month: | June |
| Day: | 24 |
| Bibtex Type: | Article (article) |
| DOI: | 10.1063/5.0092568 |
| State: | Published |
| URL: | https://aip.scitation.org/doi/10.1063/5.0092568 |
| Electronic Archiving: | grant_archive |
BibTex
@article{2022Ida,
title = {Diffusion mechanisms of DNA in agarose gels: NMR Studies and Monte Carlo Simulations},
journal = {The Journal of Chemical Physics},
abstract = {We report on the diffusion mechanism of short, single-stranded DNA molecules with up to 100 nucleobases in agarose gels with concentrations of up to 2.0% with the aim to characterize the DNA–agarose interaction. The diffusion coefficients were measured directly, i.e., without any model assumptions, by pulsed field gradient nuclear magnetic resonance (PFG-NMR). We find that the diffusion coefficient decreases, as expected, with an increase in both DNA strand length and gel concentration. In addition, we performed Monte Carlo simulations of particle diffusion in a model network of polymer chains, considering our experimental conditions. Together, the Monte Carlo simulations and the PFG-NMR results show that the decrease in diffusion coefficients in the presence of the agarose gel is due to a temporary adhesion of the DNA molecules to the surface of gel fibers. The average adhesion time to a given gel fiber increases with the length of the DNA strands but is independent of the number of gel fibers. The corresponding magnitude of the binding enthalpies of DNA strands to gel fibers indicates that a mixture of van der Waals interactions and hydrogen bonding contributes to the decreased diffusion of DNA in agarose gels.},
volume = {156},
number = {24},
pages = {245103},
month = jun,
year = {2022},
slug = {2022ida},
author = {Bochert, Ida and G{\"u}nther, Jan-Philipp and Fischer, Peer and Majer, G{\"u}nter},
url = {https://aip.scitation.org/doi/10.1063/5.0092568},
month_numeric = {6}
}