New Architectures for Long-Range Reasoning
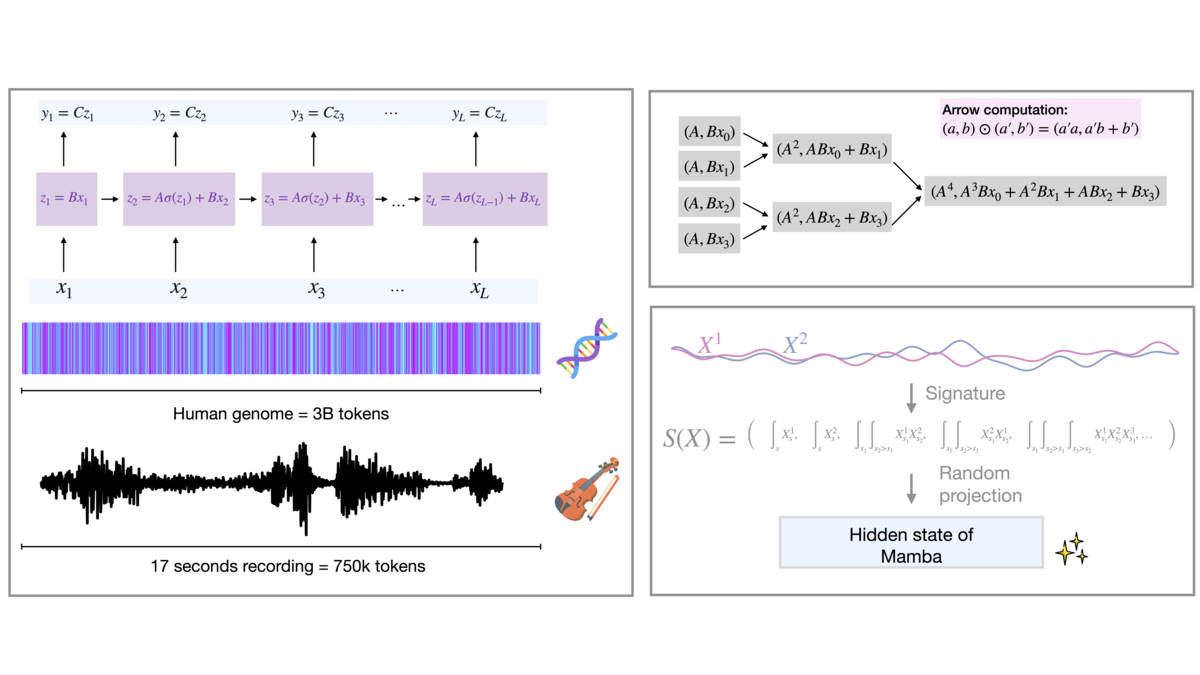
Efficiently processing long sequences is a pressing challenge in deep learning, especially in the presence of long-range dependencies. Traditional transformer models struggle with scalability due to their quadratic computational and memory demands as sequence lengths grow. State-space models (SSMs) offer a promising alternative, providing linear complexity and enabling efficient handling of longer sequences. This project seeks to advance the field by focusing on accelerating the pretraining and fine-tuning of foundation models built on SSMs and refined linear attention mechanisms. Core goals include understanding the trade-offs between compute, memory, and performance, exploring robust quantization strategies, and enabling efficient deployment of these models to extend their reasoning capabilities beyond text.
Dr. Antonio Orvieto, leading the group, has a background in optimization theory and is an active member of the community working on long-range reasoning capabilities of new foundation models - a topic of high importance for applications such as biology, language modeling, and music generation. His LRU model [], developed before joining MPI, powers the fastest architecture in Google's Gemma family. We are still actively working on the LRU architecture, as well as on similar SSM blocks (e.g., Mamba, S4, xLSTM, RWKV), offering efficient processing of very long sequences. In particular, we developed new efficient CUDA technology capable of powering these new architectures and new mathematical tools for understanding their power and limitations [
]. On top of applications to 3D vision [
] and graph data [
] we place particular attention to the biological domain. For instance, in the context of genomics, identifying functional versus nonfunctional non-coding regions of the genome has long been a challenge, with debates over which criteria—biochemical, evolutionary, or genetic—are most appropriate. AI-based knowledge discovery offers a transformative new approach, allowing the exploration of novel structures and correlations in DNA data. While recent advances in biological data availability and computational power make this a promising frontier, adapting deep learning models to DNA remains underexplored. Many recent breakthroughs in DNA analysis, such as HyenaDNA and Caduceus, are rooted in architectures designed for text rather than explicitly tailored for biological data. This highlights a gap in interdisciplinary collaboration between the biological and deep learning communities, which we are aiming to resolve with a dedicated effort.